
Hello, steemit! I am back after a long gap. :) [No No... my absence has nothing to with STEEM price. :P]
So what I am going to do today is to take you through an all-atom classical molecular dynamics exercise. But there is a difference this time. We are going to do a bigger system with a longer time scale on a High-Performance Computing(HPC) facility. The system is a DMPC lipid bilayer membrane in water with some ions. In case you want to see what a DMPC lipid is, please see my article on phospholipids linked below:
And if you want to see how I created the system of the lipid bilayer(in water) for MD experiment see my articles linked below:
- Classical Molecular Dynamics Series [Part - 4a]: Let us set up a simulation and run it!
- Classical Molecular Dynamics Series [Part - 4b]: Running small systems on your computer
System
- 288 DMPC lipids(144 in upper and lower leaflets each)
- 7776 water molecules
- 12 Sodium and 12 Chloride ions
TOTAL: 57336 atoms
The system was generated using the steps described here using CHARMM-gui tool. - Time step used = 2 femtoseconds, Total Runtime=320 nanoseconds
THE HPC FACILITY
I am going to use the HPC facility in my institute for simulating this system. Specification of the HPC is as follows:
- Nodes: 128
- Cores: 128x8=1024
- Processor : 2 x Intel Xeon Processor X5570 2.93 GHz, 6.40 GT/s Quad Core)
To run jobs on this cluster, you will need to set up a Portable Batch System (PBS) file. It is a shell script which will be capable to launch the program you need to run. I am giving a sample PBS script below. IT uses 32 nodes each with 8cores, which means total 256 cores. DMPC_patch is the job name. workq is the name of the queue. Once you login to your machine and set up the NAMD config file, you need to just run qsub pbs_script_file.sh. That will submit the below script to the job queue. The config file will run with 256 cores. Please see here for more details on PBS scripting. the most important line below is mpirun -v -f $NODEFILE /opt/app/namd/2.9/namd2 your_config_file >& your_log_file. This command transfers our namd configfile to run on 256 cores in a parallel fashion.
#!/bin/bash
#PBS -l nodes=32:ncpus=8
#PBS -N DMPC_patch
#PBS -q workq
#PBS -V
#PBS -M [email protected]
#PBS -m abe
#PBS -j oe
cd $PBS_O_WORKDIR
######Create the machinefile according the nodes allocated###############
nodes=`cat $PBS_NODEFILE`
for n in $nodes; do
echo -e ",$n 8\c" >> tmp.nodes
done
cat tmp.nodes |cut -c2- > PBS_NODEFILE_$PBS_JOBID
rm -rf tmp.nodes
#############Your application command line#########################
export MPI_DSM_DISTRIBUTE=1
NODEFILE=PBS_NODEFILE_$PBS_JOBID
mpirun -v -f $NODEFILE /opt/app/namd/2.9/namd2 your_config_file >& your_log_file
You can download the configfile from here.
How long it took for me to complete this simulation?
With the above specifications and the system size, it took around 18 days!!!. Below is a line from my output log: Info: Initial time: 256 CPUs 0.00966712 s/step 0.055944 days/ns 206.238 MB memory
.jpg)
This is the video of 320 nanoseconds long molecular dynamics simulation of a DMPC lipid bilayer patch in a water box at 290K. Water molecules and hydrogen atoms are not shown for clarity. Credits: Me :) [CLICK ON THE IMAGE TO GO TO THE DTUBE VIDEO]
WHAT HAPPENS TO DMPC MEMBRANE AT 290K?
In one of my phospholipid article, I explained about transition temperature of DMPC lipids. See the figure below:
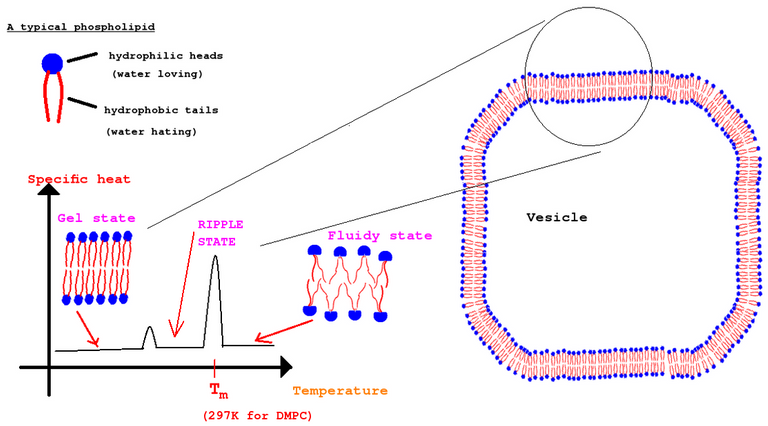
Below 297K which is the transition temperature for DMPC lipids, the membrane exhibits ordered gel phase /coexistence of gel and fluid states(Ripple state). This simulation which I performed was to see if the membrane exhibits Ripple/gel phase. If you see the video or the gif animation above you will see that towards the end of the simulation there are a lot of ordered lipids beginning to form. Initially, the lipids were like below:

And at the end of the simulation(there are many ordered lipid molecules in this case):

Another aspect of the lipids in the figure above is that lot of lipids are in interdigitated mode. By interdigitation, we mean that the top and bottom lipid legs overlap. To quantify interdigitation, we use C314 atom of extreme from DMPC tail of upper leaf layer and lower leaf layer as the selection. Then subtract the average coordinates in the z-direction. We can use interdigitation as a measure to see the phase transition of lipid from the fluid state to gel state in time. Below is the anatomy of a single phospholipid(showing C314 atom).

VMD script to measure interdigitation
mol new /home/devanandt/Documents/STEEMIT/SIMULATION_DMPC_cool/DMPC_noh_290K.psf
mol addfile /home/devanandt/Documents/STEEMIT/SIMULATION_DMPC_cool/DMPC_noh_290K.dcd first 0 last -1 step 1 waitfor all
#loading PSF and DCD file
set outfile [open avg_interdigitation_Zthick_290K.dat "w"]
#create output data file
set sel1 [atomselect 0 "name C314 and resid 1 to 144"]
set sel2 [atomselect 0 "name C314 and resid 145 to 288"]
#selections
set nf [molinfo 0 get numframes]
#count total frames
source /home/devanandt/Documents/NOGO66/ANALYSIS/MEMBRANE_ANALYSIS/average.tcl
#load tcl script to find the average of list
for {set frame 0} {$frame < $nf} {incr frame} {
$sel1 frame $frame
$sel1 update
$sel2 frame $frame
$sel2 update
set upper [average [$sel1 get z]]
set lower [average [$sel2 get z]]
set Zdiff [expr $upper - $lower]
puts $outfile "$frame $Zdiff"
#write out data to file
}
exit
Interdigitation plot (did using GNUPLOT)

CONCLUSIONS AND POSSIBLE FUTURE EXPERIMENTS
- We ran a big system on HPC facility.
- We see that when the system is cooled, it goes to a more ordered regime. This is intuitive and verifies that our simulation is correct.
- We quantified the order in the system in terms of interdigitation. This is not a perfect thing to do though, because gel phase can also exist without interdigitation. So keep this caveat in mind. We will do more analysis in future. But this simple interdigitation measurement shows us the phase transition of lipid in real-time!
- A possible future experiment is to compare this system with the heated system. So I will do a DMPC system with higher temperature and compare its interdigitation plot.
FILES (Codes and simulation related)
All necessary files available in Github repository: https://github.com/dexterdev/STEEMIT/tree/master/DMPC_290K
References [Books for further reading]
Textbook references for learning theory of Molecular Dynamics:
- "Statistical Mechanics: Theory and Molecular Simulations" by Mark E. Tuckerman
- "Molecular Modelling: Principles and Applications" by Andrew R. Leach
- "Computer Simulation of Liquids" by D. J. Tildesley and M.P. Allen
Regarding Phospholipids and biophysics etc
- Physical Biology of Cell by Rob Philips et al.. [This is a great book.]
- From biological membranes to biomimetic model membranes
References specific to NAMD and VMD:
CHARMM-gui related papers:
Join #steemSTEM

Follow me @dexterdev
____ _______ ______ _________ ____ ______
/ _ / __\ \//__ __/ __/ __/ _ / __/ \ |\
| | \| \ \ / / \ | \ | \/| | \| \ | | //
| |_/| /_ / \ | | | /_| | |_/| /_| \//
\____\____/__/\\ \_/ \____\_/\_\____\____\__/

credit: @mathowl
This post has been voted on by the steemstem curation team and voting trail.
There is more to SteemSTEM than just writing posts, check here for some more tips on being a community member. You can also join our discord here to get to know the rest of the community!
Congratulations! This excellent post was chosen by the new curation initiative of the @postpromoter content promotion service to receive a free upvote!
This post exemplifies the type of great content that we at @postpromoter enjoy reading and would love to see more of on the Steem platform. Keep up the good work!
welcome back @dexterdev
Welcome back bro. We missed your presence a lot! Hope you will be posting frequently.