Repeated growth and selection of genetically diverse populations re-domesticates brew yeast
Growth, selection, growth and more selection. This virtuous cycle of trait-guided promotion of elite plants, animals or microbes underpins the domesticated landscape that surrounds us all.
We all know that when it comes to brewing yeasts, repeated use and selection of best isolates, have led to the emergence of domesticated strains. These strains carry and use an optimised array of genetic instructions to grow and convert the wort into beer effectively.
While we all agree that domestication has happened, we know very little about the successive steps required for adaptation. How do wild yeast strains adapt to the sort of environment that wort (or beer) represents? How do yeast strains become more resistant to ethanol (or don't they?). Unfortunately, the genomes of yeast strains that already are domesticated can only give us some insight.
In a groundbreaking new study (not peer-reviewed as of yet), researchers have tried to address the question of domestication by repeating the process over six months. Specifically, the group led by Prof. Cubillos assembled a diverse population of Saccharomyces eubayanus and assessed it adaptive and evolutionary potential in the laboratory.
In a critical first step, the authors established the extent to which the thirty S. eubayanus strains used in this work can tolerate ethanol. Thirty strains were grown in 0%, 8%, 9% and 10% ethanol, to measure the impact on growth rates. These experiments revealed that ethanol, at all levels, broadly impacted the growth rate. All strains experienced reduced growth rates due to the presence of alcohol.
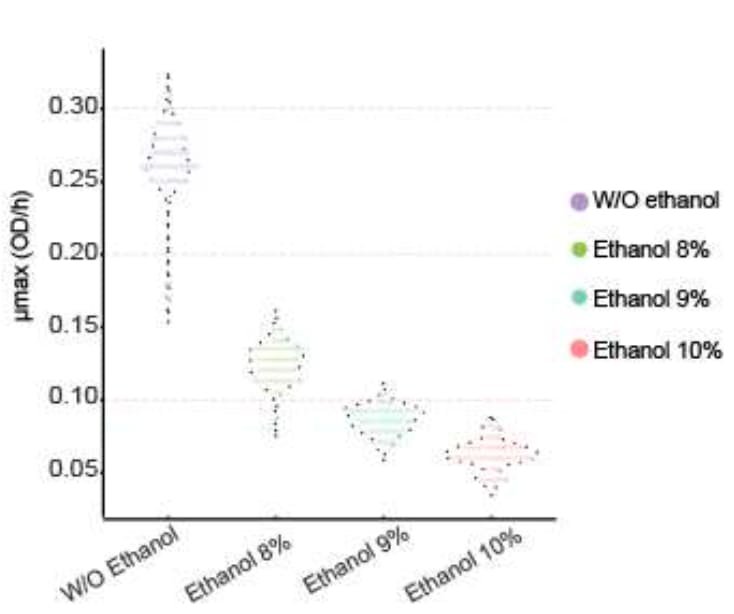
Figure 1b from Mardones et al. Growth rates (Y-axis) were estimated in varying levels of ethanol (X-axis). The presence of ethanol slowed growth rates for all strains.
The question then was: Can successive propagation of these yeast strains increase tolerance and if so, how?
To answer these questions, the authors used the thirty wildtype strains in growth and selection experiments. For this purpose, the team pooled all isolates to start ten replicate cultures (lines) in wort with ethanol and ten in the control treatment (no ethanol added). The researchers then subcultured these cultures to allow the cells to go through ~ 260 generations.
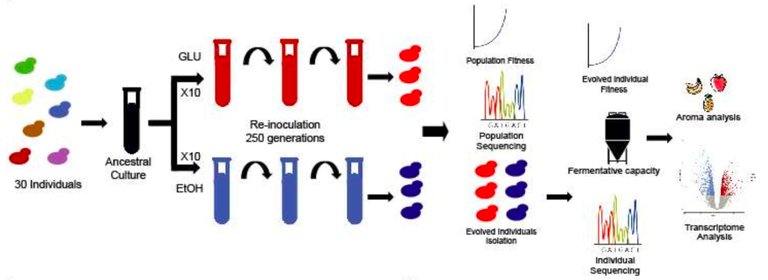
Figure 1A from Mardones et al. Schematic of yeast culturing and in vivo evolution in the presence of ethanol.
After the experiment, the authors tried to answer the following main questions:
- Are there any particular strains that can dominate the population as a consequence of ethanol selection?
- Have strains, grown in ethanol, become more resistant or tolerant?
- If strains have become more tolerant, what is the molecular and evolutionary basis of this new resistance to ethanol?
To answer the first question, the authors started to track the population make-up as cells went through successive generations. Sequencing and estimation of population composition revealed that in cultures grown in ethanol, one strain, CL248.1, dominated proceedings. Notably and in the media without ethanol, none of the strains dominated the population consistently.
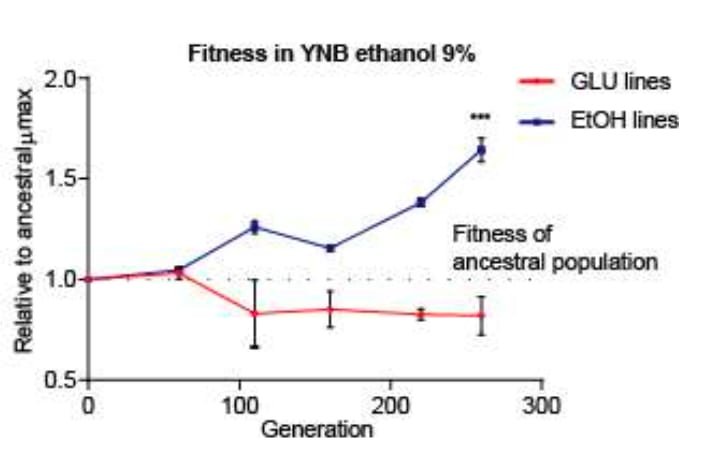
Figure 1C from Mardones et al. Propagation of yeast in media supplemented with ethanol, enhances the ability to grow in high ethanol conditions.
To answer the second question, researchers tested the growth rates of CL248.1 against other evolved strains, as well as the original (non-evolved) isolates. These assays revealed that (i) successive growth of strains in ethanol made them more resistant (based on growth rates) and (ii) that CL248.1 was not the fastest growing strain, suggestive of other factors that affected competitiveness.
To investigate the molecular mechanisms underlying enhanced alcohol tolerance, the authors used sequenced ethanol-derived lines to assess genetic diversity.
Sequence data derived from a genetically diverse population will, per definition, show this diversity. By comparing all the sequence data from the culture to a reference, you can estimate the level of diversity on the gene level.
When you measure genic (or allele) diversity over time, it generates a nice picture of how diverse the initial culture was AND how the presence of selection pressure (ethanol) affects this diversity. Consistent with a drop in population richness, you can see a decrease in genetic diversity (allele frequency). As strains (and their genes) gets eliminated from the pool, the diversity increases.
So far, the results paint a nice picture of how selective pressure can weed out a genetically diverse population. What then, is the meaning of residual genetic diversity? Does the residual genetic variation have any purpose?
The short answer is probably "yes". Remember that evolved strains have enhanced growth rates? The genetic diversity observed in thirty-four genes may specify ethanol resistance and other characteristics that allow faster growth or competitiveness. Interestingly, some of the genes identified encode factors responsible for sugar uptake, metabolism and fermentation. These observations give credence to the idea that ethanol or fermentation conditions select genes (and thereby strains) suited to deal with these conditions.
Selection does not necessarily mean genome evolution. To demonstrate an impact of long-term ethanol exposure to a particular strain's genome, you need to sequence and compare genomes before and after treatment.
This is what the authors did.
What they found is that successive growth of wildtype strains lead to significant changes to the yeast genome. Those changes included point mutations, deletions, insertions and large chromosome duplications.
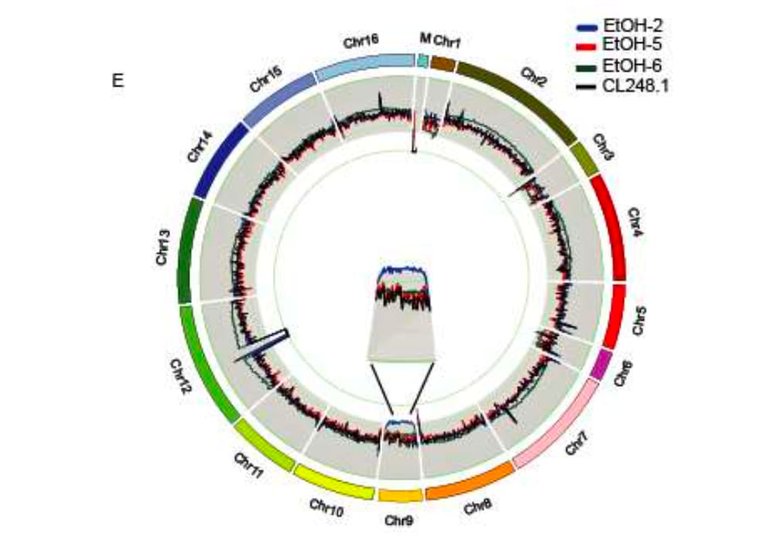
Figure 2D from Mardones et al. A whole-genome overview of changes in sequence and organisation of evolved genomes when compared to the reference genome (CL2481.)
What does that all mean?
In essence, it means that when you isolate alcohol-producing wild yeasts that are not necessarily resistant, you can grow and select for more resistant types by conducting selective growth experiments in your lab!.
There is much more to this story. If you want to learn more about this work, watch this space. The authors have kindly agreed to answer some of my questions!
Best wishes,
Edgar, The Beerologist.
Thanks for your contribution to the STEMsocial community. Feel free to join us on discord to get to know the rest of us!
Please consider supporting our funding proposal, approving our witness (@stem.witness) or delegating to the @stemsocial account (for some ROI).
Please consider using the STEMsocial app app and including @stemsocial as a beneficiary to get a stronger support.