When collecting a sample of DNA there is often a very small amount to play with. This can be very problematic for many biochemists for two main reasons.
Firstly if you are carrying out an experiment on the DNA (perhaps sequencing it, or inserting a small sequence of DNA) and it goes wrong, which can often happen with these techniques, you are in trouble. All the DNA needed for your experiment has been used up in a failed attempt! Secondly, by extracting DNA direct from the sample, it will contain all the DNA of the organism. More often than not, specific sections of DNA are required. Having large lengths of DNA can therefore make this process cumbersome and clumsy, decreasing the efficiency and increasing the possibility of error.
How are these problems overcome?
Using the Polymerase Chain Reaction (PCR) of course!
This is a staple biochemical technique which is commonly performed on small amounts of DNA before any experimentation is undertaken. PCR is able to amplify an experimenters desired segment of DNA exponentially, thus overcoming the two problems outlined above.
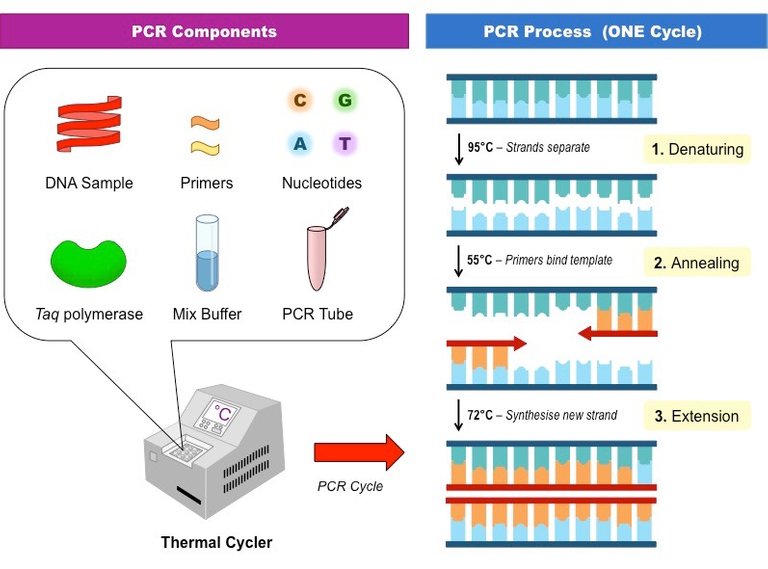
figure 1. Requirements for PCR
So, how does PCR work?
PCR proceeds in a series of cycles, and each cycle has three steps.
- Denaturation at 95°C = high temperatures separate the DNA strands
- Annealing at 55° C = lower temperatures allow the primers to bind to the template DNA
- Extension at 72°C = the DNA polymerase and nucleotides can bind to synthesise a new DNA strand.
Each cycle doubles the number of DNA strands. The process is repeated over many cycles, which causes an exponential increase in the number of DNA strands.
All this scientific jargon can become very confusing, so I am going to breakdown the roles of each of the requirements of PCR.
Primers = specific sequences of DNA around 20 nucleotdies long. They all the DNA polymerase (in this case Taq polymerase) to bind to DNA and create a new strand. Primers are required for the specificity of PCR because they will only bind to their specific complementary base sequence.
Taq polymerase = a type of enzyme known as DNA polymerase, which initially binds to the primer, then incorporates the correct nucleotides into the new DNA strand. It is called Taq polymerase because it is taken from an organism called Thermus aquaticus. It is a special form of DNA polymerase that remains active at the high temperatures during PCR (most other DNA polymerases would be denatured).
But how does PCR amplify a specific segment of DNA?
This all down to the primers.
DNA has a polarity, meaning it has a 5’ end and a 3’ end. DNA polymerase enzymes are only able to synthesise new DNA strands in one direction (5’ to 3’). Since DNA strands run in opposite directions the primers that attach either side of the target region result in DNA polymerase working in opposite directions. As shown in figure 2 below.
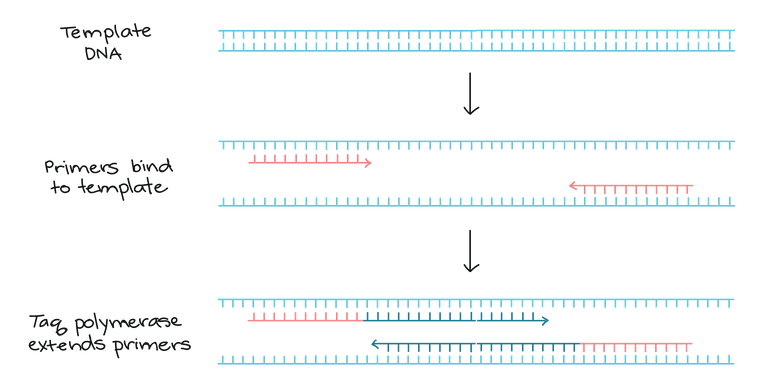
figure 2. Amplification of template DNA
This means that by repeating the PCR reaction of many cycles, only the region between the red primers will be exponentially amplified (the darker blue lines in figure 2). This is demonstrated in figure 3 below.
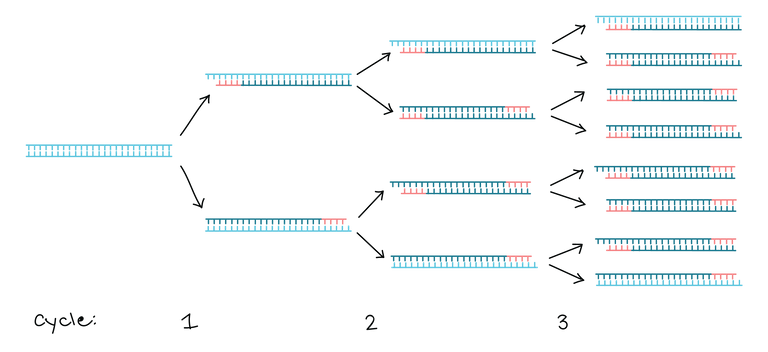
figure 3. First 3 cycles of PCR
Background reading
How would your genome be sequenced
What is DNA? Why do we need it?
References:
http://ib.bioninja.com.au/standard-level/topic-3-genetics/35-genetic-modification-and/pcr.html
https://www.khanacademy.org/science/biology/biotech-dna-technology/dna-sequencing-pcr-electrophoresis/a/polymerase-chain-reaction-pcr
https://en.wikipedia.org/wiki/Polymerase_chain_reaction
https://steemit.com/@mahboobfsd/feed
Join us on #steemSTEM / Follow our curation trail on Streemian
Thank you for this very interesting article. It has been advertised on our chat channel (and upvoted).
The steemSTEM project is a community-supported project aiming to increase the quality and the visibility of STEM (STEM is the acronym for Science, Technology, Engineering and Mathematics) articles on Steemit.
Thank you so much. I am glad that you found it interesting, and I hope to continue writing many more!
Another great informative post @ovij !! I love reading your educational posts!
Thank you. It is good to hear you are enjoying them.
Congratulations @ovij! You have completed some achievement on Steemit and have been rewarded with new badge(s) :
Click on any badge to view your own Board of Honor on SteemitBoard.
For more information about SteemitBoard, click here
If you no longer want to receive notifications, reply to this comment with the word
STOPVery nice! Follow me, I am a molecular neurobiologist who specialized in qRT-PCR, and the genetics/proteomics of learning and memory. :)
Thanks, I will definitely have a look and follow you.
Congratulations @ovij! You have completed some achievement on Steemit and have been rewarded with new badge(s) :
Click on any badge to view your own Board of Honor on SteemitBoard.
For more information about SteemitBoard, click here
If you no longer want to receive notifications, reply to this comment with the word
STOPGreat article. thanks for sharing this. Upvoted
Very interesting post. Thank you for this.