Hello and happy new year! Let us brush in 2019 by taking a read through a publication from Nature Scientific Reports titled "Molecular comparison of Neanderthal and Modern Human adenylosuccinate lyase". An article where the authors perform some biochemical and biophysical characterization of the enzyme adenylosuccinate lyase from both humans and neanderthals to gain a bit of increased understanding about how mutations can affect its activity.
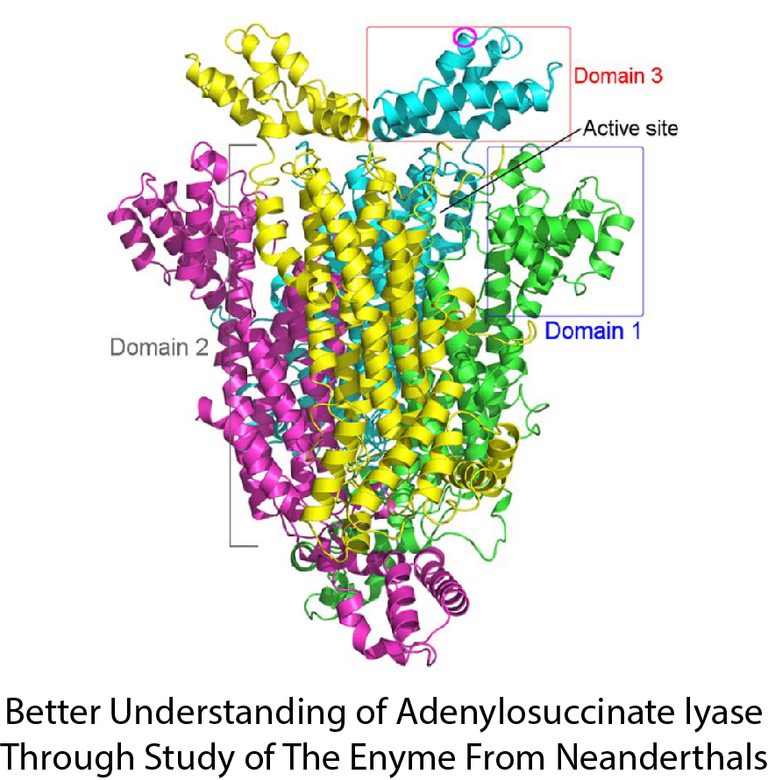
Image Reproduced From 1 Figure 2
Adenylosuccinate Lyase
Adenylosuccinate,
Reproduced From Wikipedia; Public Domain
Let us begin with a brief discussion of what this enzyme is and does. Adenylosuccinate lyase (lets call it ASL) is a enzyme that catalyzes the conversion of adenylosuccinate to adenosine monophosphate (AMP, the precursor to ATP our cellular energy currency unit), resulting in the release of a molecule of fumarate as well. Mutations can arise in this enzyme which reduce its catalytic potential, which in turn is a cause of the descriptively named disease adenylosuccinate lyase deficiency. This disease is characterized by muscle loss, movement difficulties, and even epilepsy. 2 It falls under the classification of a rare genetic disease as the population is fewer than 1 in 2000 people. 3 It is also autosomal recessive, meaning that you need two mutant genes to end up with the disease. Two parents could be carriers for the necessary mutations while not displaying the disease phenotype, however have a child who is the unlucky recipient of both diseased genes and end up with the disease.
The kinetic mechanism by which ASL functions is well understood, however the mechanisms by which the mutations alter the enzyme's ability to work are not. Thus research, as we discuss here today, is of interest to those who strive to find novel cures for these genetic derived diseases and improve the quality of life for the people that suffer with them.
Why Neanderthals?
A good question, to which there are a few answers as described by the authors.
Recent advances in DNA sequencing technology have allowed for effective reconstitution of the genomes of these highly related but still different hominids. Meaning... neanderthals are very very similar to modern Homo-sapiens, however there are still some significant differences, especially at the protein level.
Mutations to ASL result in the above described rare genetic disease
The ASL protein from neanderthals has a specific, conservative, amino acid substitution (valine 429 in the human protein is an alanine in the neanderthal version of the protein, these are two very similar amino acids... small... uncharged). Furthermore mutations in this region of the protein are known to cause disease. HOWEVER, this amino acid in the human structure is solvent exposed (its not making contacts with the rest of the enzyme, and is likely not important for its structural integrity nor enzyme activity). Will the neanderthalic enzyme function normally? Or will these predictions based on the available structural information not pan out?
Could this tell us something about how other mutations to this region of the protein DO result in disease? Could we then figure out a unique way to go after it?
So the authors set out and recombinantly expressed and purified both the human and neanderthalic version of this protein from E. coli. Then put those purified recombinant proteins through the biochemical wringer and tried to gain some understanding, from the molecular level about their differences.
Biophysical Characterization
When one goes about studying the effects that mutations have on a protein, one thing that can be quite informative is to look for changes in protein stability. One such way to see if there is a change is to use a technique called thermal denaturation, where you slowly heat a sample of protein and watch for changes in spectroscopic readings. In this article the authors were using a technique I regularly do, protein thermal melting with an orange dye called 'Sypro Orange'.
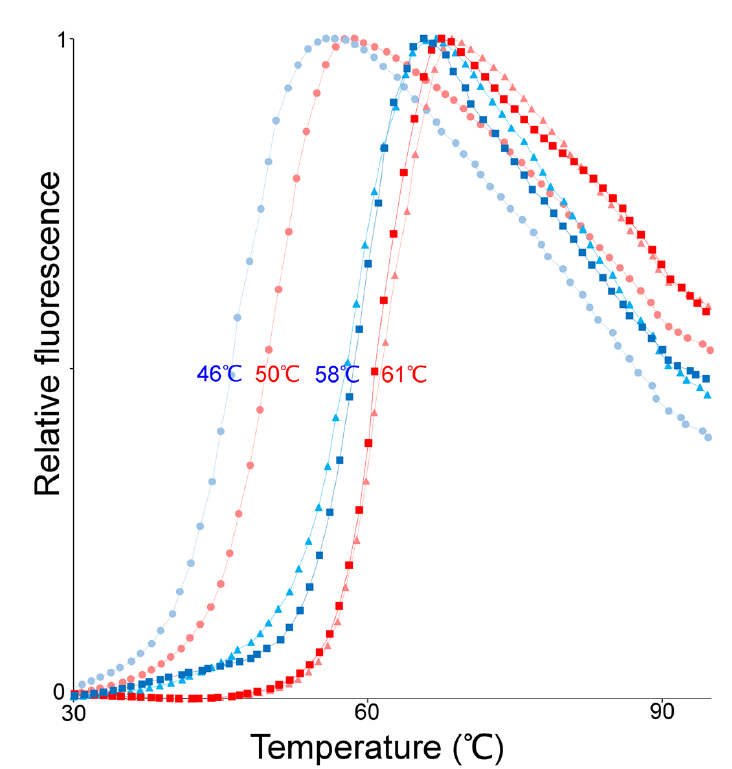 Reproduced from [1] Figure 1
Reproduced from [1] Figure 1 Sypro is used because it glows more brightly when it is bound to something hydrophobic, like regions inside the core of a globular protein. As the protein sample is heated the protein blob begins to unfold, this exposes those hydrophobic regions to the dye which then binds. As more and more of the protein unfolds more and more dye binds and the fluorescence gets brighter and brighter. Thus we have a really nice probe for monitoring the protein unfolding process.
In the figure to the right (and perhaps above depending on the size of your browser window) we are looking at the results of the thermal unfolding, fluorescence starts low and begins to increase as temperature is increased (you get these nice S shaped curves). The researchers used a few different conditions so there are multiple curve sets (squares, triangles, circles) for both proteins (human ASL is in blue, neanderthal ASL is in red) lets not worry about the conditions but focus on red vs. blue. What we see is that for every set, the red curve unfolds at a slightly higher temperature than the blue curve (there is more specific information we can derive from this sort of data set but I don't think you want me to teach you the full ins and outs of a thermal unfolding experiment). This means that the neanderthal protein is actually MORE stable (in this thermal denaturation assay at least) than the modern human protein. Wait, wasn't this mutation supposed to be conservative and play no structural role?
Brief on crystallographic analysis: The authors also crystallized the neanderthal ASL protein and used X-Ray crystallographic techniques to determine the structure of the protein. They then compared that structure to that of the human enzyme (which had already long been known). They observed that the change of the valine to the alanine in the neanderthalic enzyme resulted in no significant structural changes. The authors also looked for differences in the enzymes when bound to their product (AMP and Fumarate) and saw no significant differences there between the two proteins either. So no observable structural change... but... a thermostability change. Hmm.
In their crystallographic analysis the authors also observed that both enzymes undergo a change in their structures upon binding their product. Thus they wanted to explore whether or not there was a conformational change which is associated with their catalytic function. They orthogonality (used a second method) confirmed using a technique called small angle X-ray scattering or SAXS that both proteins underwent a conformational change (IE there is movement in the protein structure that coincides with the reaction that the enzyme catalyzes).
Additional Biochemistry
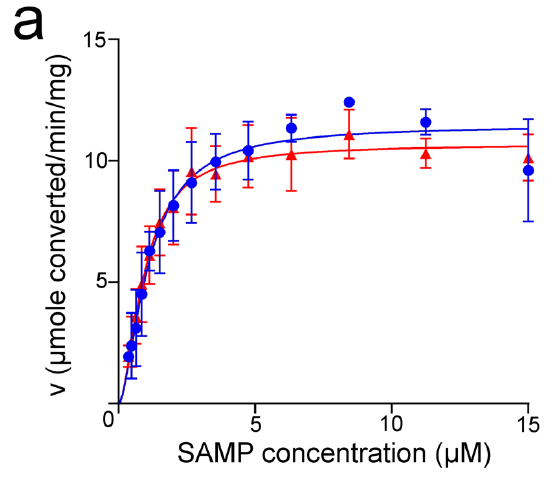 Reproduced From [1] Figure 5A
Reproduced From [1] Figure 5AThe authors then proceeded to do some enzyme kinetics (shown in the figure to the left) where they looked at the conversion of Adenylosuccinate (SAMP) into AMP by monitoring the absorbance of a reaction solution containing various concentrations of the SAMP substrate in the presence of the enzyme and watched as the absorbance at 282 nM decreased as SAMP was consumed by the enzyme. They then plotted the rate of each reaction against that concentration of SAMP substrate (seen in the figure) to determine some kinetic parameters for the two enzymes.
However here, unlike the thermostability what they saw is that both the forward reaction (what I am showing above) and the reverse reaction (I am not showing) occurred with similar kinetic parameters for both the human (blue) and neanderthalic (red) enzymes.
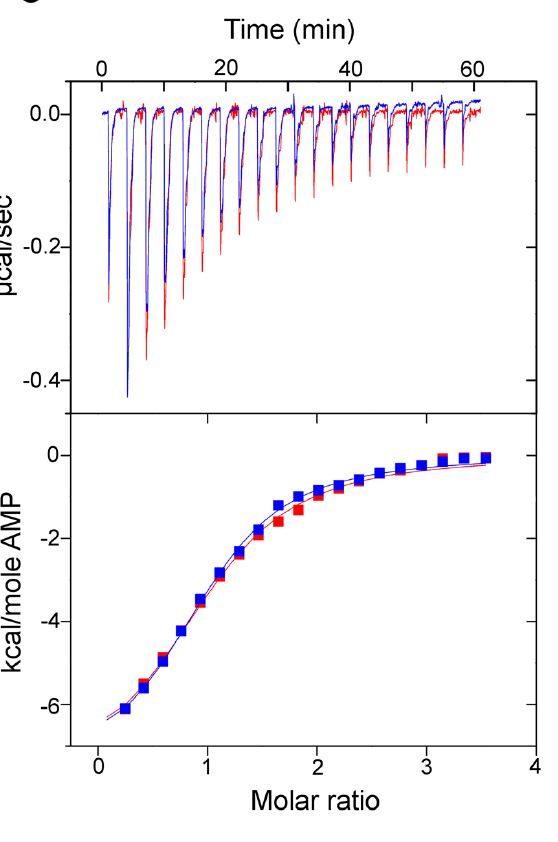 Reproduced from [1] Figure 5C
Reproduced from [1] Figure 5CThey also looked into whether or not there were any differences in substrate binding affinity through a technique called isothermal titration calorimetry (ITC) which monitors the microscopic amount of heat that gets released as the substrate molecule binds to the protein. However, similarly (and not at all unsurprising considering the enzyme kinetic results) the substrate binding of both enzymes is very very similar (the ITC data between the two overlap!).
So this change in sequence that we observe from neanderthals to humans, despite occurring in a similar location does not result in significant structural or biochemical perturbation of the enzyme. Do the mutations which cause human disease behave similarly or are these different? (based on prior literature they should have reduced catalytic activity and perturbed stability)
To look at this the authors made proteins harboring three disease causing mutations in the human protein sequence. (R396C, N422T, and R426H) They then went back and re-visited the enzyme kinetics and thermal unfolding experiments.
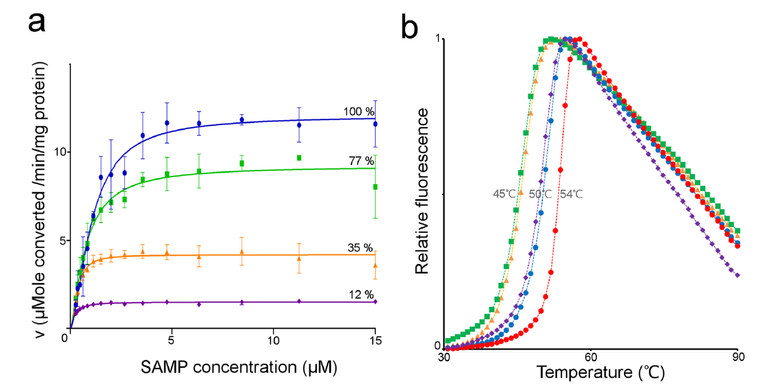
Reproduced from 1 Figure 5
Here we see that unlike with the wild type human, or neanderthalic enzymes, that the three disease causing mutations (to varying degrees) result in a both reduced enzyme thermostability as well as a change in maximal enzyme rate (IE decreased enzyme activity). In the figures the Blue trace is the wild type human enzyme, the green is the human enzyme harboring the R426H mutation, orange is N422T and the purple is R396C. What is interesting to me is that the thermostability results do not look to correlate with the enzyme activity results. For instance look at the purple trace (R396C mutation) there we see that the enzyme is severely reduced in its activity (just 12% of wild type level) however it doesn't really look much different from a thermal unfolding perspective (purple and blue traces overlap). However the green is most perturbed from the thermostability perspective (it has the lowest unfolding temperatures) but is not all that reduced in enzyme activity.
What we understand from this is that a disease phenotype can be caused both from a mutation which drastically perturbs enzyme activity, but also from one which does not. Which is important as we now know that we can not make assumptions based entirely on the activity of the enzyme (which is something that we enzymologists.... love to do :D )
Summary of Findings
This is a fairly well studied enzyme, however in this study the authors revealed that contrary to prior thoughts, a conformational change does occur during the enzymes catalytic functioning. They also show that the conservative mutation harbored by neanderthals, A429V, does not cause any change in enzyme catalytic function unlike the disease causing R426H (just 3 amino acid residues away) does. And while not expected to cause a change in thermostability actually results in an enzyme which is more thermostable.
The data here illustrates that genetic predictions revolving around enzyme activity alone are not sufficient to predict disease causing mutations. Rather, it is only through the combination of multiple biological, biochemical, and biophysical disciplines that we can gain sufficient information to truly begin piecing the puzzle together about how genetic alterations result in disease.
Cited Materials
- https://www.nature.com/articles/s41598-018-36195-5
- https://doi.org/10.1023/A:1005323512982
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4029002/
Make sure to follow steemstem on steemstem.io, steemit, facebook, twitter, and instagram to always be up-to-date on our latest news and ideas.
Please also consider to support the project by delegating to @steemstem for a ROI of 65% of our curation rewards (quick delegation links: 50SP | 100SP | 500SP | 1000SP | 5000SP | 10000SP).
Follow us || Vote for the SteemSTEM Witness || Visit our new home steemstem.io
The Neanderthal part left me speechless... So, we have access to their genome, don't we. That sounds pretty cool to me!
PS: I think there are more fancy words in your fields than in mine ;)
Their complete genome yes. It's not all that different from ours. But then we share 50% sequence homology with a banana so ... All life is surprisingly similar.
My field is specifically the study of enzyme function. All of the genetics is the level above me primarily :)
All of the fancy words, someone else gets to do those studies and reconstructions... :(
I guess what matters is not the common part but the difference, isn't it?
Correct
Is this the first analysis of a Neanderthal protein? If so I'm guessing that's the main draw of the paper. Cool find either way.
I am not sure if it is the very first, but clearly it was the draw of the paper. Their data was a whole lotta not much though. (If it were I kinda suspect a higher impact journal than Scientific Reports)
It seems to me like their isn't a large body of good mechanistic biochemistry on this enzyme. So most working knowledge is based purely on biology and genetic context.
I agree with you. Though you could still trace the genome of the Neanderthals (Homo sapien neanderthalensis) to some of the extant modern humans (Homo sapiens sapiens). And not just that, even the Denisovan genomes are present in some people.
And this leaves me with this question; how do we differentiate the pure breed of Homo sapiens sapiens from the hybrid? Because from inference; it looks like there were some cross-breedings that happened behind the scene.
I'm not a Biochemist though; I'm only speaking from a paleoanthropological point of view.
Nice piece sir
Computational Genomics. Though I am not specialized in that. There were cross breeding's but only in specific subsets of the population of mondern humans. I don't imagine it's that hard to identify a consensus sequence for the average homosapien gene vs neanderthal.
This is a very nicely written article. I honestly love your writing style. There's real work to be done by scientists in finding cures to these rare genetic diseases. And that's why I couldn't agree with you more when you say
Thanks a lot for sharing!
Hey thanks for reading it!
Yes, there are many of us (me included) hard at work trying to give the people suffering with these diseases some sort of treatment, to restore at least some semblance of quality of life.
Each of these diseases posts their own unique challenges, and the amount of work is massive. I am hopeful that technologies like CRISPR can play a role in helping deal with these genetic issues, but at least to my eyes that tech is further off than many would like. At least in any meaningful wide spread use sort of capacity.
Onward we press!
Fluorescence of proteins, my first love <3 and M.Sc. thesis, although I never used SYPRO Orange (shame on me)
Sypro isn't a protein fluorescent tag. Its just a free dye which has a propensity to bind to anything hydrophobic. I can stick any greasy thing in the buffer and the fluorescence will spike. In my work, I deal with a lot of small molecules, and some of which can cause some really wonky data due to background interactions between the dye and the compound. Obscuring the protein unfolding that I am interested in.
We liked doing it Raw :D Tryptophan only
Those were very boring experiments:
Now I'm returning back to fluorescence and fancy probes for STED
When you use internal fluorescence you typically need to use a lot of protein 10-20 uM. However with sypro you can get very nice unfolding curves, usually with very minimal purturbation of intrinsic unfolding Tm (based on Trp fluorescence), at concentrations in the 1 - 2 uM range, and I have gotten down to as low as 250 nM for a few proteins. Depends on the amount of hydrophobic surface.
These are still boring experiments, I typically multiplex and do things in 384 well plate using a Q-PCR machine. So you can generate a whole bunch of data all at once.
Another fun little path is to use the 'proteoplex' software to quantitate and optimize unfolding conditions, really speeds up assay design. (http://www.proteoplex.de/)
This post made me miss being a enzymologist.
Would you like me to make more content like this? I didn't realize there was interest in actual biochemical discussion. I was under the impression that people preferred biological context. However if there is interest, I'd be happy to talk more about the stuff I actually understand.
Yes, the biochemist in me really appreciates it.
Very interesting piece! Thanks for sharing.
Glad you thought so! Thanks for giving it a read.
The adjective 'rare' in these cases always has me shaking my head. It seems pretty frequent to me. Plus, almost all diseases are either as 'rare' as that or less rare, and I'm pretty sure there's more than 2000 genetic diseases, which means we all suffer from something.
And that doesn't sound rare to me :(
Everyone gets colds, or the flu, and you get them multiple times. 9.4% of the population of the Us has diabetes. 4:1000 get diagnosed with cancer (every year!), by contrast these genetic diseases are at birth and in comparison to the aformentioned diseases are 'rare.'
Doesn't mean people don't suffer with them. It also doesn't mean we aren't working for cures. :)
I have always thought that X-ray crystallography is one of the most complex and difficult techniques to perform, so I value a lot this kind of work, very interesting and illustrative, greetings.
Thanks for reading through! Cheers.
Hi @justtryme90!
Your post was upvoted by Utopian.io in cooperation with @steemstem - supporting knowledge, innovation and technological advancement on the Steem Blockchain.
Contribute to Open Source with utopian.io
Learn how to contribute on our website and join the new open source economy.
Want to chat? Join the Utopian Community on Discord https://discord.gg/h52nFrV
Hi @justtryme90!
Your post was upvoted by @steem-ua, new Steem dApp, using UserAuthority for algorithmic post curation!
Your UA account score is currently 6.356 which ranks you at #192 across all Steem accounts.
Your rank has dropped 1 places in the last three days (old rank 191).
In our last Algorithmic Curation Round, consisting of 351 contributions, your post is ranked at #38.
Evaluation of your UA score:
Feel free to join our @steem-ua Discord server
Congratulations @justtryme90! You have completed the following achievement on the Steem blockchain and have been rewarded with new badge(s) :
Click here to view your Board
If you no longer want to receive notifications, reply to this comment with the word
STOP