Point mutation in protein-coding genes can affect protein structures in a variety of ways. Point mutations can be named according to if and how they effectively change the encoded protein. Silent mutation, missense mutation, nonsense mutation and frameshift mutation are types of point mutations.
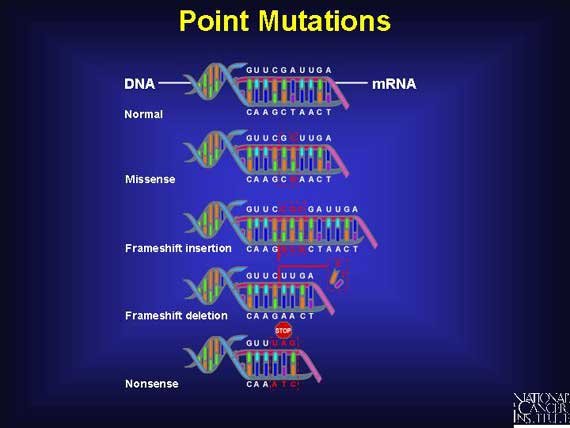
Image Source: Wikimedia Public Domain
Silent Mutation
This type of mutation changes the nucleotide sequence of a codon but do not change the amino acid encoded by that codon. This is made possible because the genetic code exhibits degeneracy. So, whenever there is more than one codon for a given amino acid, a single base substitution may lead to the formation of a new codon for the same amino acid. Let's take for instance, if the codon CGU were changed to CGC, it would still code for arginine, even as the mutation has occurred. Whenever there is no change in the protein, there will also be no change in the phenotype of the organisms.
Missense Mutation
This involve a single base substitution that changes a codon for one specific amino acid into a codon for another. For instance, the codon GAG which specifies glutamic acid could be change to GUG, which codes for valine.
Effects of missense mutation may vary, they alter the primary structure of a protein, but the effect of this change may vary from complete loss of activity or no change at all. This is simply because the effect of missense mutation on protein function is dependent on the type and position of the amino acid substitution. For example, replacing a non polar amino acid in the protein's interior with a polar amino acid can severely alter the 3D structure of the protein and therefore its function as a protein structure is a unique determinant of its function. Similarly, the replacement of a critical amino acid at the catalytic site of an enzyme often destroys it's activity, however, the substitution of one polar amino acid with another polar amino acid at the protein surface may have little or ni effect at all. Such mutations are termed Neutral Mutation.
Missense mutation plays a very important role in providing new variability to drive evolution as they are often not lethal and therefore remain in the gene pool.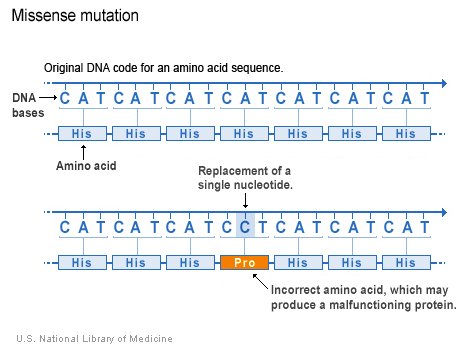
Image Source: Wikimedia Public Domain
Nonsense Mutation
This type of mutation converts a sense codon (One that codes for amino acid) to a nonsense codon (A stop codon: One that does not code for amino acid). This leads to an early termination of translation and therefore causes a shortened polypeptide. Depending on the position of the mutation in the gene, the phenotype may be more or less drastically affected. Most protein retain some of their function if they are shortened by only one or two amino acids; complete loss of normal function usually occurs if the mutation happens closer to the beginning or middle of the gene.
Frameshift Mutation
This type of mutation arises from the addition or deletion of base pair that is within the coding region of the gene. In as much as the code consist of a precise sequence of three codon, the addition or deletion of less than three base pair causes shifting of the reading frame to be shifted for all other subsequent codon downstairs. Frameshift mutation produce mutant phenotypes resulting from the synthesis of non functional proteins. However, frameshift mutation often produce a stop codon so as to make the peptide product shorter as well as different in sequence. Of course, if the frameshift happens near the end of the gene or if there is a second frameshift shortly downstream from the first that restores the reading frame, the phenotypic effect might not be severe.
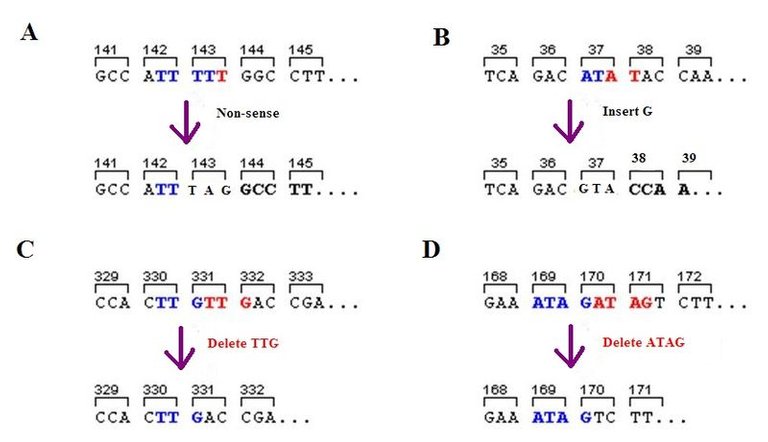
Image Source: Wikimedia CC BY-SA 3.0
e severe.
Conclusion
As learned from above, changes in protein structure can alter the phenotype of an organism in many ways.
Thanks for reading through and upvotes , comments and resteems will appreciated.
References